Chat with Molecule Neo (Gemini)
An advanced AI-powered assistant for MoleditPy that enables intelligent, conversational interaction with molecular structures using Google’s Gemini API.
Key Features
- Deep Context Awareness: Automatically injects SMILES strings, atom map numbers, and user selections into the AI’s reasoning engine.
- AI-Driven Structural Tools: The assistant can autonomously modify molecules, highlight functional groups, and generate complex QM input files.
- PubChem Integration: Seamlessly identifies molecules and fetches chemical data via the PubChem API.
- Visual Chemistry: Native LaTeX rendering for chemical formulas and mathematical equations using Matplotlib.
- Interactive Links: Molecular names are formatted as clickable
smiles:links that load structures directly into the editor.
AI Tool Suite
The AI assistant has direct access to the MoleditPy engine through a suite of 11 specialized tools:
| Tool | Capability |
|---|---|
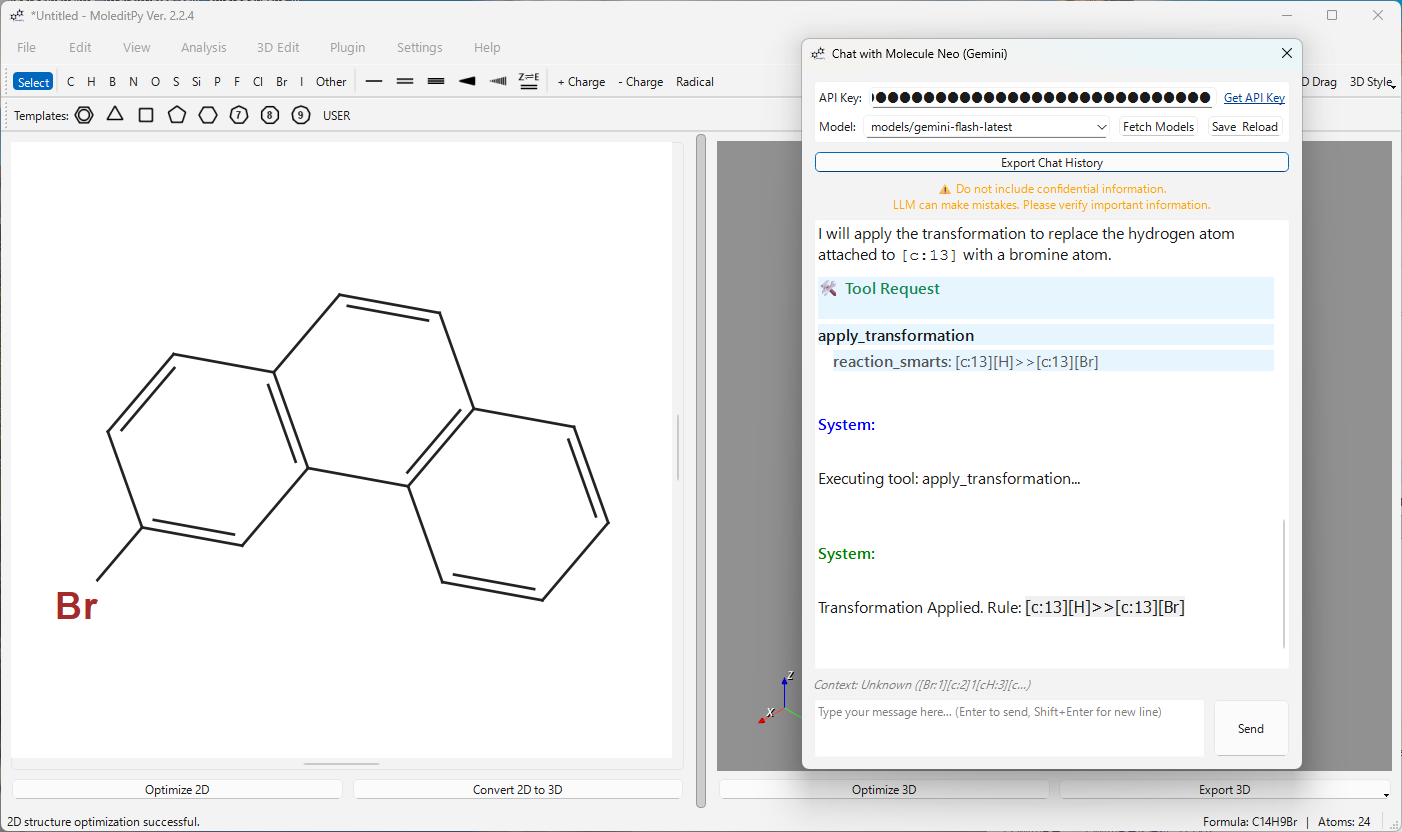
| Apply Transformation | Perform chemical reactions via SMARTS patterns (e.g., chlorination, methylation). |
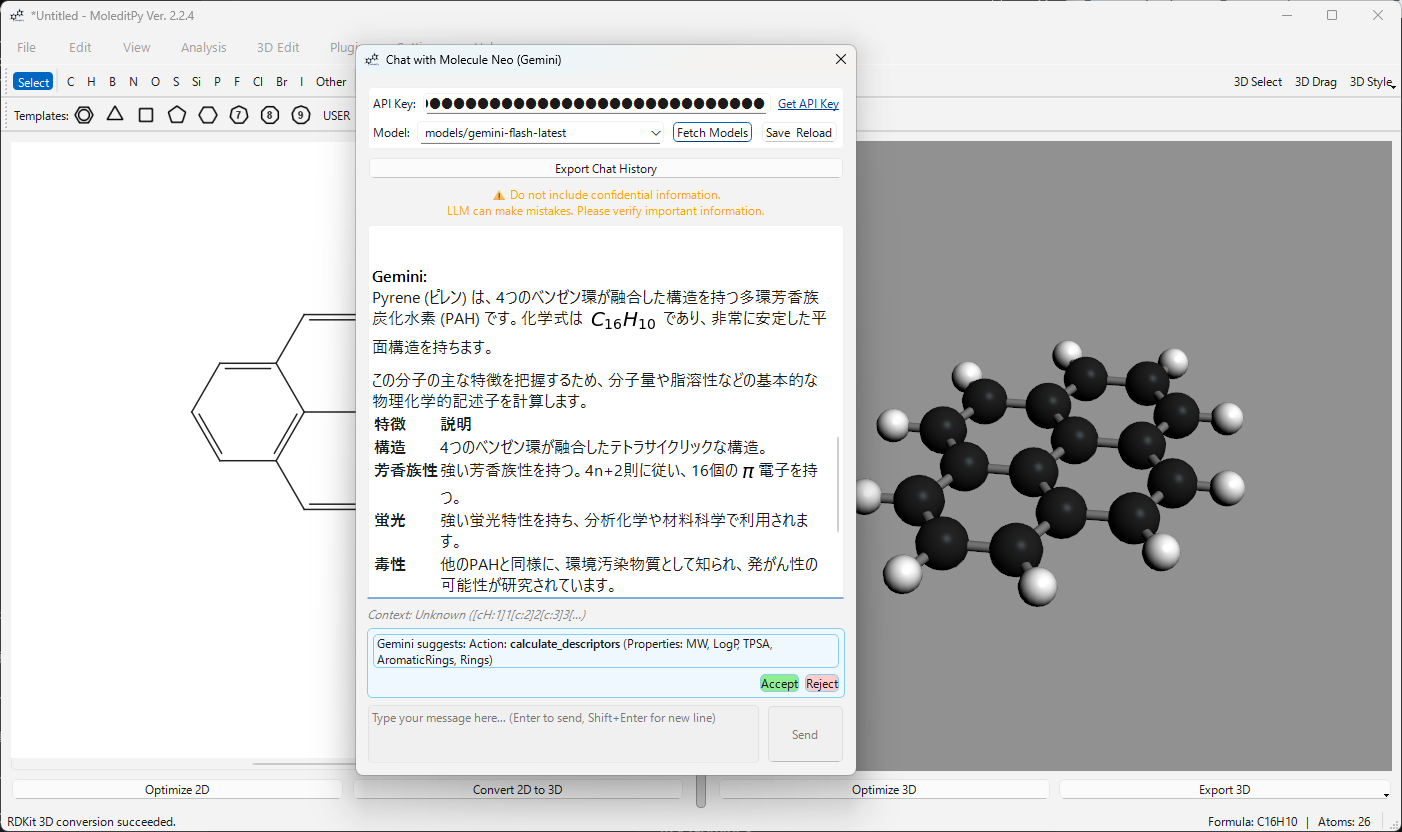
| Calculate Descriptors | Get MW, LogP, TPSA, HBD/HBA, and ring counts instantly via RDKit. |
| ORCA Input Generator | Create production-ready ORCA (.inp) input files with coordinates and headers. |
| Gaussian Input Generator | Create production-ready Gaussian (.gjf) input files with coordinates and headers. |
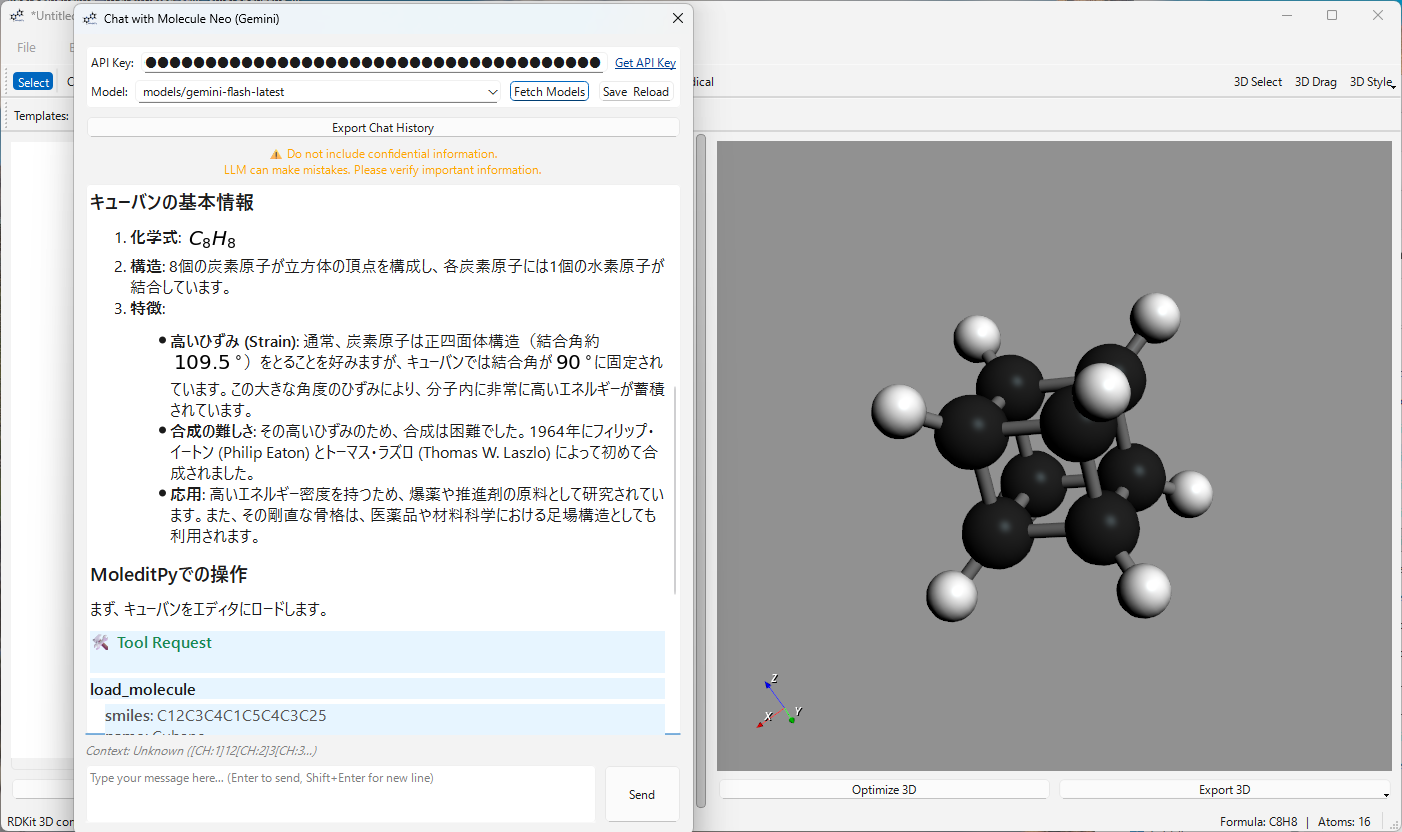
| Load Molecule | Directly load any molecular structure into the editor using a SMILES string. |
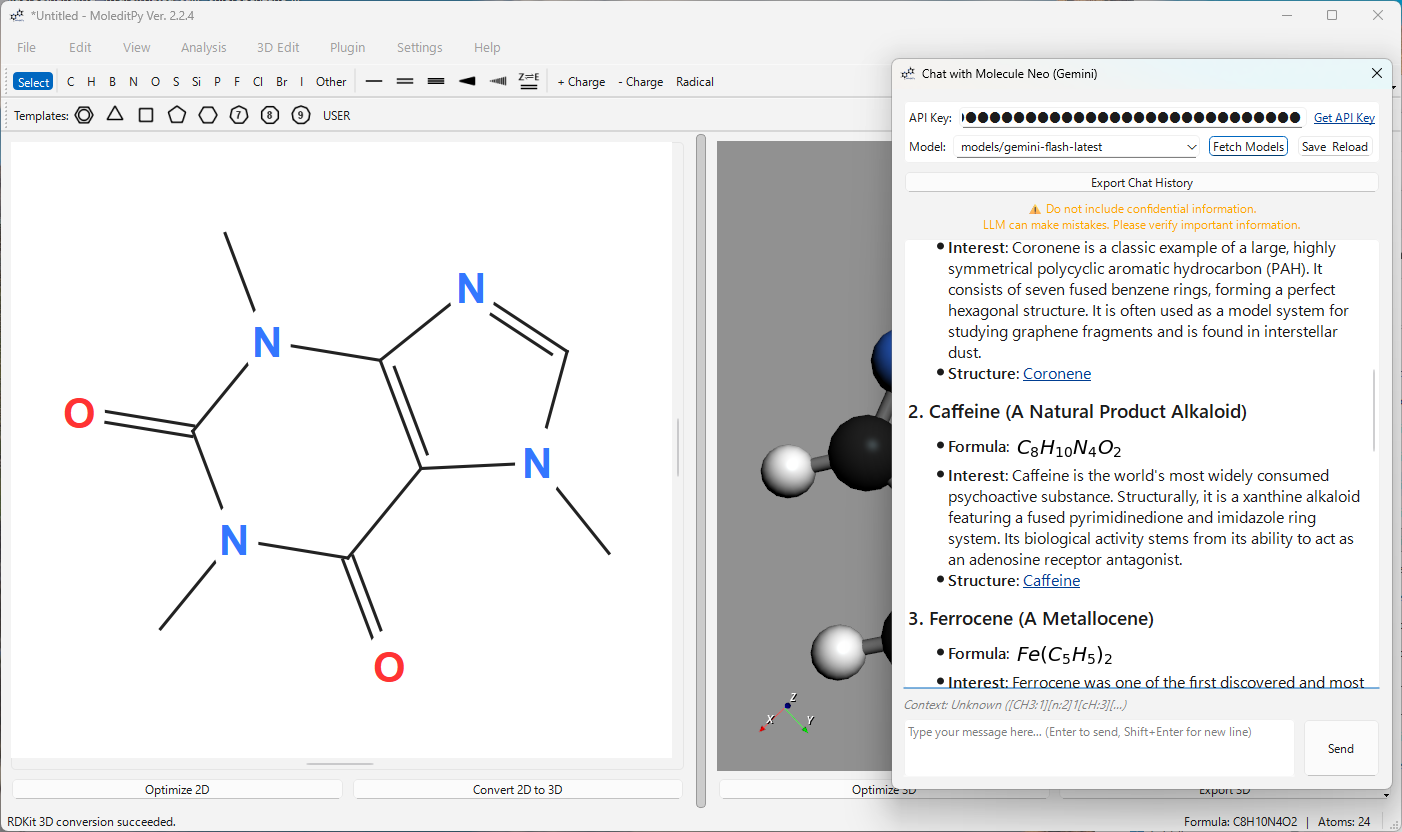
| Load Molecule by Name | Search PubChem by name (e.g., “Aspirin”, “Caffeine”) and load the structure automatically. |
| Highlight Substructure | Visually identify functional groups or specific atom indices in the 3D viewport. |
| Set Electronic State | Precisely adjust formal charges and multiplicities for specific atoms. |
| Convert to 3D | Trigger MMFF94/UFF force-field optimizations and 3D visualization. |
| Clear Canvas | Reset the current workspace for a fresh start. |
| Save Generic Files | Generate and save scripts, CSVs, or custom coordinate files (XYZ, etc.). |
Core Chemistry Toolkits
This plugin leverages industry-standard computational chemistry libraries:
- RDKit: The primary engine for cheminformatics, handling SMILES parsing, SMARTS matching, coordinate generation, and property calculations.
- PubChem API: Used for asynchronous chemical name resolution and structural identification via InChIKeys.
- Matplotlib (Agg): Provides the rendering engine for dynamic LaTeX-based math and chemical formula visualization in the chat UI.
Installation
This plugin requires Python 3.10+ and the following dependencies:
pip install google-generativeai markdown matplotlib rdkit
Setup
- Get an API Key: Obtain a free API key from Google AI Studio.
- Configuration:
- Open MoleditPy.
- Navigate to Plugins > Chat with Molecule Neo.
- Paste your key into the API Key field.
- Select your preferred model (e.g.,
gemini-pro-latestorgemini-flash-latest). - Click Save & Reload.
Usage Examples
1. Molecular Modification
User: “Add a nitro group to the para-position of the benzene ring.” AI: Analyzes the graph, identifies the para-carbon, and applies the transformation.
2. Quantum Chemistry Setup
User: “Prepare an ORCA input for a geometry optimization using B3LYP/def2-SVP.” AI: Generates the
.inpfile with correct atom coordinates and header directives.
3. Property Analysis
User: “Calculate the Lipinski’s Rule of Five parameters for this molecule.” AI: Invokes the descriptor calculator and provides a detailed summary.
4. Load by Name (PubChem Search)
User: “Load Caffeine” or “アスピリンをロードして” AI: Searches PubChem, retrieves the SMILES, and loads the structure into the editor.
Aesthetics & UI
- Streaming Responses: Real-time feedback as the AI generates its explanation.
- Safety First: Built-in verification for tool execution and clear warnings for AI-generated content.
Technical Architecture
Chat with Molecule Neo is built as a robust, non-blocking plugin using the following stack:
- GUI Framework: Built with PyQt6, inheriting from
QDialogto provide a secondary interactive window. - Asynchronous Processing:
GenAIWorker (QThread): Handles streaming responses from the Google Gemini API to prevent UI hangs.InitWorker: Manages secure API configuration and model discovery.QThreadPool: Executes PubChem name resolution and property fetching in the background.
- Chemistry Engine: Leverages RDKit for molecular graph analysis, SMILES generation, and SMARTS-based reaction execution.
- Secure Navigation: Custom
ChatBrowsersubclass disables internal navigation to prevent page-clearing errors, handlingsmiles:andhttp:schemes viaQDesktopServices.
Integration Mechanics
The plugin deeply integrates with the MoleditPy core:
- State Synchronization: Uses a 2-second polling mechanism (or focus-triggered checks) to monitor
main_window.data.atomsfor structural changes or selection updates. - Context Injection: Automatically constructs a system context including the current molecule’s SMILES, InChIKey (resolved via PubChem), and specific atom selection indices.
- Tool Execution Protocol: The AI outputs JSON-formatted commands which are parsed and executed by the plugin against the live MoleditPy
MainWindowinstance, allowing for real-time canvas updates.
Universal Disclaimer for AI Features (Cloud & Local)
Disclaimer regarding AI Integration
This software integrates with external AI services (e.g., OpenAI API, Google Gemini API) and local LLM backends (e.g., Ollama). By using these features, you acknowledge and agree to the following:
- No Warranty on Accuracy: The content generated by AI models—regardless of whether they are cloud-based or running locally—may contain errors, hallucinations, or scientifically inaccurate information. The developer makes no warranty regarding the correctness or safety of the generated chemical structures or explanations. Users must independently verify all data.
- Cloud Services (OpenAI / Google, etc.):
- BYOK Policy: Users are responsible for managing their own API keys and securing them.
- Costs & Compliance: Users are solely responsible for any usage fees incurred and for compliance with the respective provider’s Terms of Service. The developer is not liable for account suspensions or financial charges.
- Local LLMs:
- Hardware Usage: Running local models requires significant computational resources. The developer is not responsible for system instability, overheating, or performance degradation of the user’s hardware.
- Model Performance: Local models may have different capabilities compared to cloud models. The user assumes full responsibility for selecting appropriate models for their tasks.
- Limitation of Liability: The developer shall not be held liable for any direct or indirect damages arising from the use of these AI features.
Author
HiroYokoyama Feel free to contribute or report issues via the Plugin Explorer.

